Part of a series of articles titled Yellowstone Science - Volume 27 Issue 1: Vital Signs - Monitoring Yellowstone's Ecosystem Health.
Article
SHORT: The Yellowstone River Fish-Kill

The Yellowstone River Fish-Kill: Fish Health Informs and Is Informed by Vital Signs Monitoring
by Patrick R. Hutchins, Adam J. Sepulveda, Lacey R. Hopper, & Ken D. Staigmiller
Trout are socioeconomically and ecologically important in the Greater Yellowstone Area (GYA), yet these fish face numerous threats. Disease may begin to play a larger role in reducing fish populations, partly because many existing threats may interact to exacerbate the frequency, extent, and severity of fish diseases (Lafferty 2009). For example, habitat loss and low summer flows might interact to stress fish, making them more susceptible to disease while also increasing fish densities in microhabitats, thereby creating conditions where infectious diseases are more easily spread. Conservation and management efforts to mitigate these threats often involve reactionary measures to unforeseen events. Long-term monitoring of aquatic vital signs and fish health, however, may provide important insights for predicting the spread of fish diseases and the extent and severity of outbreaks.
A need for monitoring the health of wild fish populations was first recognized in 1996 after the causative agent for whirling disease, Myxobolus cerebralis, was identified in Montana's Madison River. This parasite decimated wild trout populations and prompted the U.S. Fish and Wildlife Service (USFWS) to request funding for the National Wild Fish Health Survey (NWFHS), which aims to detect known pathogens and to discover new or emerging pathogens that may threaten fish populations. With changing environmental conditions and high potential for movements of pathogens from endemic to naive waters, continuous monitoring of wild fish health may be a necessary component of a comprehensive vital sign monitoring campaign.
The 2016 mass mortality event in the Yellowstone River highlighted the complexities of fish disease. Between mid-August and mid-September thousands of dead mountain whitefish (Prosopium williamsoni) were documented between Emigrant and Springdale, Montana (Opitz and Rhoten 2017). In response to the event, Montana Fish, Wildlife and Parks (FWP) closed 183 miles (295 km) of the Yellowstone River and all tributaries to all recreational activities. The USFWS’s Bozeman Fish Health Center identified Proliferative Kidney Disease (PKD) as the cause of the mortalities. PKD is characterized by anemia and severe inflammation of the kidney (Hedrick et al. 1993). Clinical signs usually only develop when water temperatures exceed 59°F (15°C) for several weeks or longer (Ferguson 1981).
While its presence had been reported in Montana, the causative agent of PKD in salmonid fish, Tetracapusloides bryosalmonae, was not known to be present in the Yellowstone River. This raised concern of a recent introduction of the parasite to the Yellowstone River and wrought fear of its spread upstream towards Yellowstone National Park. In the wake of this outbreak, many questions loom, including: How long has the parasite been in the Yellowstone River? Are extreme environmental conditions to blame for the 2016 PKD outbreak? Why were no outbreaks of that magnitude observed in neighboring water bodies known to harbor the parasite? Are mountain whitefish, in particular, more susceptible to the disease than trout?
What We Have Learned Since the 2016 PKD Fish Kill
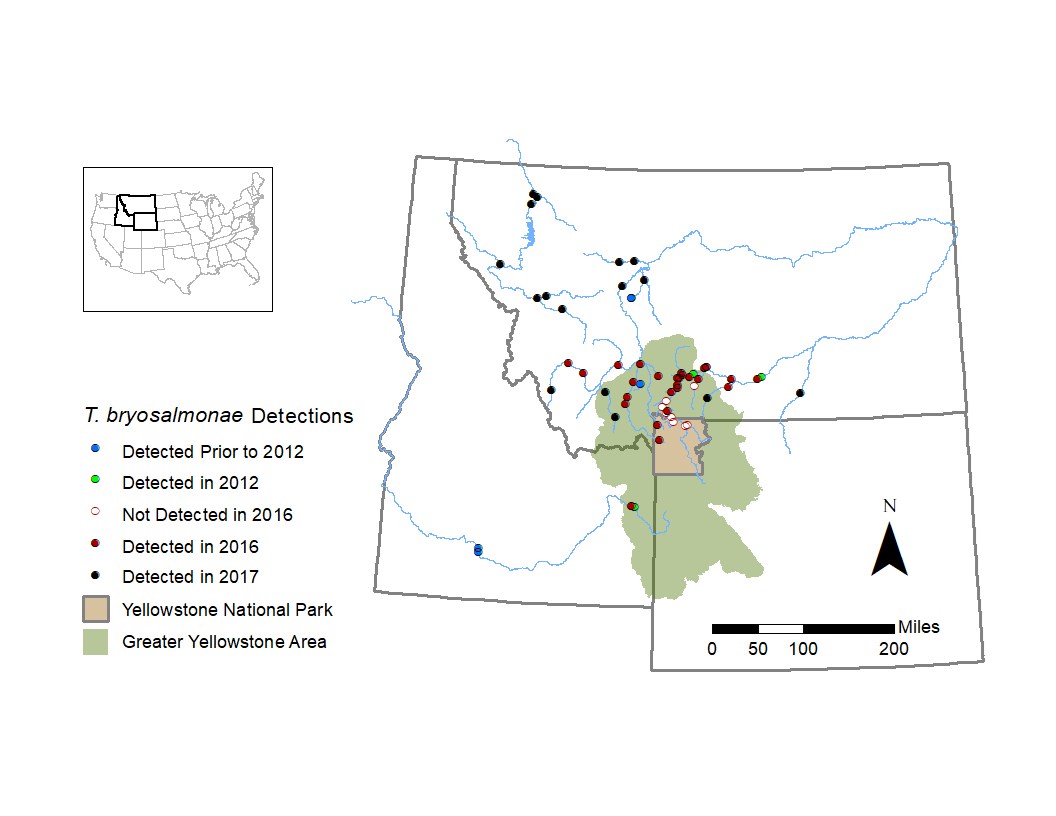
To learn about PKD and T. bryosalmonae, the U.S. Geological Survey’s (USGS) Northern Rocky Mountain Science Center has partnered with the USFWS Fish Health Center and Montana FWP to develop new molecular tools for T. bryosalmonae surveillance and to use these tools to describe the occurrence and distribution of T. bryosalmonae in rivers in the GYA and western Montana. In winter 2017, this collaborative effort developed enhanced molecular tools for screening water and fish tissue samples for presence of T. bryosalmonae DNA (Hutchins et al. 2017, 2018). These tools were then used to screen tissue samples collected from apparently healthy trout and whitefish in Montana rivers in 2016 and 2017, as well as archived fish kidneys preserved in 2012 from the Yellowstone River (figure 1). Presence of the parasite in a water sample or in fish tissue is not indicative of diseased fish or fish mortality and our findings likely do not represent the full extent of T. bryosalmonae’s distribution in the GYA or the Intermountain West. We did, however, document T. bryosalmonae DNA in nearly all waters where fish were sampled, including multiple rivers in the GYA (figure 1).
Many regional rivers had hydrologically stressful conditions similar or worse than the Yellowstone River in 2016, yet there were no documented PKD fish kills. These results indicate that T. bryosalmonae is widely distributed and suggest that warm temperatures and low flow conditions cannot alone explain PKD-caused fish kills. This unanticipated die-off further underscores our limited understanding of PKD. Focused monitoring of PKD disease dynamics is required to better understand how stressors interact to result in PKD mortality events.
A Framework for Monitoring Fish Diseases
The health of fish populations is subject to a wide-range of biotic and abiotic factors. Tracking patterns in fish disease or parasite prevalence, therefore, may serve as an integrative measure of ecosystem health not revealed though tracking physical vital signs (e.g., flow and water temperature) alone. Traditional fish health surveys typically involve lethal sampling of fish so that tissues can be diagnosed in the lab. Diagnostic approaches include visual identification, culture-based methods, and, more recently, molecular techniques.
Environmental DNA (eDNA) approaches, however, can detect target strands of DNA that are freely available in the environment. Sampling water, rather than fish tissue, can be used to noninvasively monitor natural waters for the presence of infectious diseases and with less expense and labor than traditional sampling (Huver et al. 2015). Parasite detection in water involves filtration of easily collected water samples that are then used in downstream molecular diagnostics. However, detecting parasite DNA in water does not imply the presence of a viable parasite population or indicate widespread infection in fish at that location. For this and other reasons (e.g., relative low cost), eDNA should not be used as the sole line of evidence for fish disease, but should be used as a monitoring tool that informs more intensive sampling efforts.
Surveys for parasite DNA in water can be used to inform research and management efforts at varying temporal and spatial scales. For example, eDNA samples collected at USGS streamgages can pair parasite DNA detections and environmental conditions over large temporal and spatial scales, while eDNA samples collected along transects spanning several river miles may help identify parasite “hotspots” at smaller temporal and spatial scales. Both sampling strategies can be used in isolation or in concert to inform and enhance how the NWFHS is implemented. Patterns in parasite detection can, for instance, inform targeted fish collection for tissue screening to determine the prevalence and severity of infection in sampled fish. Using eDNA, therefore, has great potential to add power, flexibility, and utility to existing fish health and vital sign monitoring programs.
Literature Cited
Ferguson, H. 1981. The effects of water temperature on the development of proliferative kidney disease in rainbow trout, Salmo gairdneri Richardson. Journal of Fish Diseases 4:175–177.
Hedrick, R., E. MacConnell, and P. De Kinkelin. 1993. Proliferative kidney disease of salmonid fish. Annual Review of Fish Diseases 3:277–290.
Hutchins, P.R., A.J. Sepulveda, R.M. Martin, and L.R. Hopper. 2017. A probe-based quantitative PCR assay for detecting Tetracapsuloides bryosalmonae in fish tissue and environmental DNA water samples. Conservation Genetics Resources. doi:10.1007/s12686-017-0812-3.
Hutchins, P.R., A.J. Sepulveda, R.M. Martin, and L.R. Hopper. 2018. Improved conventional PCR assay for detecting Tetracapsuloides bryosalmonae DNA in fish tissues. Journal of Aquatic Animal Health doi:10.1002/aah.10020
Huver, J.R., J. Koprivnikar, P.T.J. Johnson, and S. Whyard. 2015. Development and application of an eDNA method to detect and quantify a pathogenic parasite in aquatic ecosystems. Ecological Applications 25:991–1002.
Lafferty, K.D. 2009. The ecology of climate change and infectious diseases. Ecology 90:888–900.
Opitz, S., and J. Rhoten. 2017. 2016 Mountain whitefish kill on the Yellowstone River. Montana Fish, Wildlife and Parks, Bozeman, Montana, USA.
Patrick Hutchins is a Molecular Ecologist for the U.S. Geological Survey operating out of the Northern Rocky Mountain Science Center in Bozeman, Montana. He uses genetics and genomics approaches to address aquatic disease and populations issues in ecology.
Last updated: September 18, 2019
