Abstract
Despite the critical roles fungi play in the functioning of ecosystems, especially as symbionts of plants and recyclers of organic matter, their biodiversity is poorly known in high-latitude regions. Among these, Beringia, including Alaska and north-eastern Siberia, has long been a focal point for biogeographical research in a wide range of plant and animal taxa. However, the biodiversity and biogeography of fungi in Beringia are virtually unknown. We analyzed DNA sequence data from various boreal and arctic macrofungi using phylogenetic and coalescent methods to assess the genetic diversity at the species and intraspecific levels. Our results suggest that Beringia, particularly Alaska, harbors very diverse fungal communities and that most arctic and at least some boreal fungal taxa survived the last glacial maximum in Beringia.
Introduction
Climatic changes in the Quaternary have dramatically influenced the distribution of mycota, flora and fauna in high-latitude ecosystems, and had major impacts on past speciation events and present population structures. While plants and animals have been extensively studied, very little is known about the community and population structures of fungi in arctic and boreal biomes. This is particularly undesirable, because fungi play key roles in the decomposition, mobilization, and the transfer of nutrients to plants in these nutrient-poor ecosystems.
Beringia, including Alaska and north-eastern Siberia, has long been a focal point for biogeographical research in a wide range of plant and animal taxa. This high level of interest arises for two principal reasons. First, due to its diverse landscape and climate and the fact that much of the region remained ice-free during glacial maxima, Beringia served as a refugium for arctic and subarctic flora and fauna (Adams and Faure 1997, Brubaker et al. 2005, Edwards et al. 2000, Hultén 1968). Second, during much of the Tertiary and Quaternary periods, Beringia was the major land con-nection between Asia and North America and provided migration routes to a wide variety of organisms (for example, see Elias et al. 2000; Qian 1999, Swanson 2003).
As opposed to plants and animals, there has not been a comprehensive cataloging of fungi in Alaska, and the richness and biogeograhic origins of Beringia’s mycota remain unknown. Therefore, one of our primary goals was to initiate the first biodiversity assessment of boreal and arctic fungi in Alaska, con-ducting genetic analyses based on curated sporocarp collections. Here, we present an example of a genus-wide diversity assessment in the ectomycorrhizal Lactarius Pers.
Beside exploring species-level diversity, we hypothesized that, similar to patterns documented in various plants and animals (e.g., see MacNeil and Strobeck 1987, Sperling and Harrison 1994, Abbott and Comes 2003), high intraspecific genetic diversity can be found in Beringian fungi. To test it, we sampled populations of selected boreal and arctic fungi from regions across the Northern Hemisphere and carried out phylogenetic and coalescent analyses.
Methods
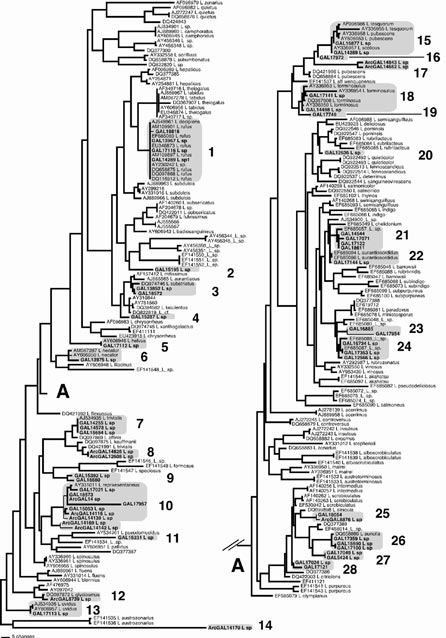
For the genus-wide diversity assessment of Lactarius, 383 specimens were collected and deposited in the Mycological Herbarium (GAL) at the University of Alaska Fairbanks (UAF). To reduce redundancy, 58 of these, representing morphological groups and geographic areas of origin among the collections, were selected for molecular work. Nucleotide sequences of the de-sired portions (ITS and LSU rDNA) of the DNA samples were obtained using standard molecular protocols (DNA extraction, polymerase chain reaction, cycle sequencing etc.). Additional sequence data of all Lactarius species available in GenBank was downloaded and in-corporated into multiple sequence alignments and phylogenetic analyses for reference. Putative Alaskan species groups were detected as phylogenetic groups of sequences.
Four species (Amanita muscaria, Lichenomphalia umbellifera, Flavocetraria cucullata, and Flavocetraria nivalis) were chosen for intraspecific analyses, based on their circumpolar distributions and the availability of materials from the major northern geographic regions. Molecular data was obtained as described above and was subjected to phylogenetic and coalescent analyses to study the population histories and characteristics.

J. Geml
Results
Phylogenetic analyses revealed 28 putative species groups in the genus Lactarius in Alaska. These were broadly distributed on the genus-wide tree and grouped with several major infrageneric groups. It was often possible to identify these groups to known species, although in many cases Alaskan sequences formed unique, unidentified clades that may represent newly discovered entities (Figure 1).
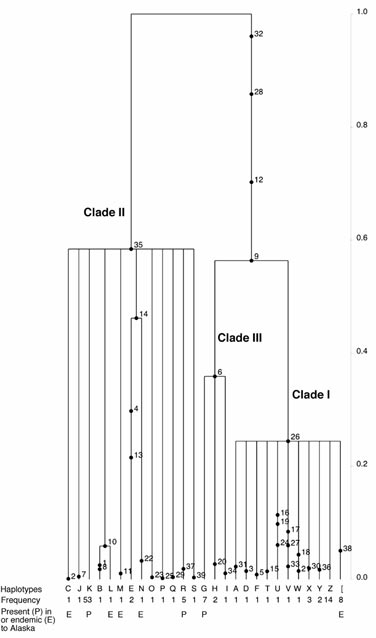
The intraspecific analyses showed very high genetic di-versity in Alaska, particularly in the boreal ectomycorrhizal mushroom Amanita muscaria. In this morphological species, we discovered three non-interbreeding phylogenetic species that occur in sympatry in Alaska (Figures 2-3). Two of these are ‘Eurasian’ clades (Clades II-III), while one is North American’ (Clade I), based on their geographic distribution. To our knowledge, the ‘Eurasian’ clades do not occur in North America outside Alaska. Furthermore, coalescent analyses revealed the genetic isolation of the endemic Alaskan sequence types from the rest of their ancestral population, suggesting local survival for an extended period, including at least one glacial maximum.
All three arctic fungi sampled in our study also showed high intraspecific diversity, but they lacked significant phylogeographic structure, likely as a result of frequent, long-distance dispersals across the Arctic. Despite the effective strategies for rapid postglacial colonization, there were slight differences among different arctic regions, Alaska hosting reliably the most diverse populations.
Discussion and conclusions
Biodiversity: Arctic and boreal plant communities are frequently described as relatively species poor and having simpler patterns than those in more southern biomes (e.g., Whittaker 1975, Scott 1995, Walker 1995). Our genus-wide diversity assessment suggests that at least one group of ectom corrhizal fungi, the genus Lactarius, likely is diverse in Alaska, particularly when comparing our data to the other estimates of basidiomycete diversity (O’Brien et al 2005, Allison et al 2007). Based on the phylogenetic breadth of our sequences, most, if not all, known major phyloge-netic groups of Lactarius are represented in Alaska. This is in sharp contrast to the trend seen in the non-mycorrhizal saprotrophic Agaricus L.:Fr., where only three section-level phylogenetic clades (half of the six known globally) are represented in Alaska (Geml et al. 2008). Our future plan is to assess the diversity of other important Alaskan genera: Russula, Cortinarius, Hebeloma, Inocybe, Galerina etc.
Putative forest refugia during the Last Glacial Maximum in Alaska: Whether fragments of boreal forest existed in Beringia during the Last Glacial Maximum (LGM) is a major, but, as yet, unanswered question in quaternary science. Because most of the discussion has been centered on palynological data, using molecular phylogeography of ectomycorrhizal fungi, as presented here, may help us better understand past vegetation patterns in Beringia. The likely importance of host trees in the distribution of ectomy corrhizal fungi has been repeatedly noted, given the obligate nature of the symbiosis, particularly from the fungal perspective.
Our data show support for at least two endemic regional populations of A. muscaria in different parts of Alaska, both of which exhibit genetic isolation and differentiation from other populations. Because non-Alaskan populations most likely survived the LGM in refugia south of the major ice shields, the lack of migration between these and the Alaskan ones suggests local survival of the latter, implying forest refugia in Alaska. Our findings support the existence of at least two independent such glacial forest refugia: 1) boreal forest in Interior Alaska; and 2) maritime rainforest in Southeast Alaska and the Pacific Northwest. The possible existence of isolated forest refugia in Interior and Southeast Alaska is also supported by several other independent lines of evidence (e.g., see Flemming and Cook 2002, Carrara et al 2003, Brubaker et al 2005, Weckworth et al 2005, Anderson et al 2006).
High intraspecific diversity and long-range dispersal in arctic fungi: Despite the high genetic diversity observed, we found no phylogeographic structure in the three arctic species examined (L. umbellifera, F. cucullata, and F. nivalis), indicating high levels of gene flow across the Arctic. Several sequence types, particularly the ancestral ones, were distributed over multiple continents, suggesting effective dispersal. As opposed to morphological species from boreal and temperate regions that often comprise multiple evolutionary lineages, morphological species and phylogenetic species seem to correspond well in the arctic fungi we analyzed. In other words, there appear to be no genetic isolation among populations inhabiting different geographic areas. On the other hand, slight differences still remain in the overall genetic diversity among different regions, and the high diversity values observed in Alaska, for example, could be explained by the glacial history and/or the climatic and landscape variability.
Management implications
We are providing pioneer data on the diversity of Alaskan fungi, including the discovery of several putatively novel species. Such baseline information is crucial for preserving biodiversity and ecosystem function in Alaska national parks. Also, the resulting ‘DNA barcode’ database is useful for current and future ecological and biodiversity studies. Finally, insights into fungal migration histories and observed common patterns contribute to improved inferences concerning glacial refugia and to the understanding of the present geographical structure of genetic diversity in arctic organisms. Knowledge of both past migration history, a key to prediction, and present day genetic diversity are essential to respond intelligently to global change.
Acknowledgements
The authors gratefully acknowledge the following sources for financial and logistical support: University of Alaska IPY Office, Humboldt Foundation, National Science Foundation, National Centre for Biosystem-atics (University of Oslo), Institute of Arctic Biology (University of Alaska Fairbanks), and National Science Foundation (grant no. 0333308: Metagenomics of Boreal Forest Fungi project). We thank colleagues who contributed specimens to our study and appreciate the help of Shawn Houston and James Long at the Biotechnology Computing Research Group (University of Alaska Fairbanks) for assistance in bioinformatics.
References
Abbott R.J., and H.P. Comes. 2003.
Evolution in the Arctic: a phylogeographic analysis of the circumarctic plant, Saxifraga oppositifolia (Purple saxifrage). New Phytologist 161: 211-224.
Adams J.M., and H. Faure. 1997.
Preliminary vegetation maps of the World since the last glacial maximum: An aid to archaeological understanding. Journal of Archeological Science 24: 623-647.
Allison, S.D., C.A. Hanson, and K.K. Treseder. 2007.
Nitrogen fertilization reduces diversity and alters community structure of active fungi in boreal ecosystems. Soil Biology and Biochemistry 39: 1878–1887.
Alsos, I.G., T. Engelskjøn, L. Gielly, P. Taberlet, and C. Brochmann. 2005.
Impact of ice ages on circumpolar molecular diversity: insights from an ecological key species. Molecular Ecology 14: 2739-2753.
Anderson, L.L., F.S. Hu, D.M. Nelson, R.J. Petit, and K.N. Paige. 2006.
Ice-age endurance: DNA evidence for a white spruce refugium in Alaska. Proceedings of the Nationa
Brubaker, L.B., P.M. Anderson, M.E. Edwards, and A.V. Lozhkin. 2005.
Beringia as a glacial refugium for boreal trees and shrubs: new perspectives from mapped pollen data. Journal of Biogeography 32: 833-848.
Carrarra, P.E., T.A. Ager, J.F. Baichtal, D.P. VanSistine. 2003.
Map of glacial limits and possible refugia in the southern Alexander Archipelago, Alaska, during the late Wisconsin Glaciation. Miscellaneous Field Studies Map, MF-2424. US Geological Service. Denver, CO.
Edwards, M.E.,P.M. Anderson,L.B. Brubake,T.A. Ager, A.A. Andreev, N. Bigelow,L.C. Cwynar,W.R. Eisner, S.P. Harrison, F.S. Hu, D. Jolly,A.V. Lozhkin, G.M. MacDonald, C.J. Mock, J.C. Ritchie, A.V. Sher, R.W. Spear,J.W. Williams,and G.Yu. 2000. Pollen-based biomes for Beringia 18,000, 6000 and 0 14C yr BP. Journal of Biogeography 27: 521-554.
Elias, S.A., D. Berman, and A. Alfimov. 2000.
Late Pleistocene beetle faunas of Beringia: where east met west. Journal of Biogeography 27: 1349-1363.
Fleming, M.A., and J.A. Cook. 2002.
Phylogeography of endemic ermine (Mustela erminea) in southeast Alaska. Molecular Ecology 11: 795-807.
Geml, J., G.A. Laursen, and D.L. Taylor. 2008.
Molecular diversity assessment of arctic and boreal Agaricus taxa. Mycologia 100(4): 577-589.
Hultén, E. 1968.
Flora of Alaska and Neighboring Territories. Stanford University Press. Stanford, CA.
MacNeil, D. and C. Strobeck. 1987.
Evolutionary relationships among colonies of Columbian ground squirrels as shown by mitochondrial DNA. Evolution 41: 873-881.
O’Brien, H., J.L. Parrent, J.A. Jackson, J.M. Moncalvo, R. Vilgalys. 2005.
Fungal community analysis by large-scale sequencing of environmental samples. Applied and Environmental Microbiology 71: 5544-5550.
Qian, H. 1999.
Floristic analysis of vascular plant genera of North America north of Mexico: characteristics of phytogeography. Journal of Biogeography 26: 1307-1321.
Scott, A.J. 1995.
Canada’s vegetation: a world perspective. McGill-Queen’s University Press. Montreal, Québec.
Sperling, F.H., and R.G. Harrison. 1994.
Mitochondrial DNA variation within and between species of the Papilio machaon group of swallowtail butterflies. Evolution 48: 408-422.
Swanson, D.K. 2003.
A comparison of taiga flora in north-eastern Russia and Alaska/Yukon. Journal of Biogeography 30: 1109-1121.
Walker, M.D. 1995.
Patterns and Causes of Arctic Plant Community Diversity. In Arctic and Alpine Biodiversity, edited by S. Chapin III and S. Körner. 3-20.
Weckworth, B.V., S. Talbot, G.K. Sage, D.K. Person, J. Cook. 2005.
A signal for independent coastal and continental histories among North American wolves. Molecular Ecology 14: 917-931
Whittaker, R.J. 1975.
Communities and Ecosystems. Macmillan. New York.
Part of a series of articles titled Alaska Park Science: Volume 8, Issue 2: Park Science in the Arctic.
Last updated: February 11, 2019
